Introduction
Sacheon City, located along the southern coast of Gyeongsangnam-do, South Korea, represents a unique convergence of diverse geographical and ecological environments within a relatively compact area, encompassing mountains, plains, coastlines, islands, rivers, and wetlands. The region exhibits a temperate oceanic climate typical of the southern coastal zone, with mild winters, hot, humid summers, and distinct seasonal transitions that support a dynamic, productive natural ecosystem. The coastline forms a complex ria-type shoreline with extensive tidal flats-features rarely observed in inland environments-creating conditions highly favorable for the development of rich marine biodiversity. Owing to these geographical and climatic characteristics, Sacheon supports a vibrant marine ecosystem and has considerable potential for advancing the marine and fishery industries, including aquaculture, marine biotechnology, and the sustainable utilization of coastal biological resources. Furthermore, the integration of coastal agriculture, moderate-scale livestock and forestry industries, and advanced marine transportation infrastructure positions Sacheon as a strategically significant region where land, sea, and air-based industries coexist synergistically (Park et al., 2025; Yang et al., 2025). Despite this ecological richness, research on the exploration and industrial utilization of microbial resources in Sacheon remains limited. The implementation of the Nagoya Protocol under the Convention on Biological Diversity has intensified the need to secure, characterize, and utilize region-specific microbial biodiversity for potential bioindustrial development. In response, the Sacheon Microbial Fermentation Foundation, established in 2020, has initiated collaborative studies with the EF-Bio Laboratory at Silla University.
Marine microorganisms have evolved under high pressure, salinity, and low-temperature conditions, producing unique secondary metabolites that differ from those produced by terrestrial microbes (Zhang & Kim, 2010). As a maritime nation surrounded by three seas, South Korea possesses diverse marine bioresources, offering significant potential for the development of novel functional and medicinal foods (Moreno et al., 2013). Traditional Korean fermented seafood products, including salted fish sauces, seaweed pickles, and seasoned seafood dishes, are prepared through autolytic and microbial enzymatic processes under high-salinity conditions, in which halophilic bacteria play a crucial role in flavor development and texture modification (Sekar & Kim, 2020). Halophilic bacteria are known producers of industrially valuable hydrolytic enzymes such as proteases, amylases, and lipases, which are widely utilized in the food, detergent, textile, leather, and pharmaceutical industries, as well as in the recycling of fishery by-products (Barzkar et al., 2020). Moreover, several halophilic strains have demonstrated the ability to synthesize auxin (e.g., indole-3-acetic acid, IAA), thereby promoting plant growth and offering promise for use as eco-friendly biofertilizers (Etesami & Glick, 2024). For example, halophilic and halotolerant bacteria isolated from coastal soils have been shown to produce IAA alongside other plant growth-promoting traits (Reang et al., 2022). In this study, halophilic bacterial strains were isolated and characterized from marine environments and seafood (including fermented products) collected in Sacheon, South Korea. The enzymatic activities (amylase, lipase, and protease) and auxin production potential of the isolates were evaluated to explore their applicability in various industrial and agricultural sectors. These findings provide a fundamental basis for securing and utilizing halophilic microbial resources from the Sacheon coastal ecosystem.
Materials and Methods
Marine environmental samples and fermented seafood products were collected from diverse habitats in the Sacheon region, South Korea, including seawater, tidal flats, coastal sediments, marine mud, and traditional homemade fermented seafoods (Table 1). To minimize contamination, sterile gloves were worn during sampling, and sterilized spatulas were used to collect each sample. The samples were transferred into sterile 50 mL conical tubes and transported to the laboratory in an ice box under chilled conditions. Upon arrival, all samples were stored at 4°C until further processing. All microbial strains isolated during this study were deposited in the Microbial Value Enhancement Program, Korea Research Institute of Bioscience and Biotechnology (KRIBB), Daejeon, South Korea.
To isolate region-specific halophilic microorganisms, sixteen samples of marine environmental materials and fermented seafood products were collected from various sites in Sacheon, South Korea. Each sample (1 g) was suspended in 9 mL of sterile 0.85% (w/v) saline solution and homogenized using a magnetic stirrer to prepare a uniform suspension. The resulting suspension (1 mL) was serially diluted tenfold from 10–1 to 10–4 using sterile saline. Aliquots of each dilution were spread onto Marine Agar (MA; BD Difco, USA), a medium formulated for the cultivation of marine microorganisms. The inoculated MA plates were incubated aerobically at 37°c. Following incubation, colonies exhibiting distinct morphological characteristics (size, shape, color, and texture) were selected and streaked repeatedly on fresh MA plates to obtain pure isolates.
To evaluate the growth adaptability, each strain was cultivated on Nutrient Agar (NA; BD Difco, USA), Reasoner’s 2A Agar (R2A; BD Difco, USA), and Tryptic Soy Agar (TSA; BD Difco, USA) at 37°c for seven days under static conditions. Salt tolerance and pH adaptability were assessed on MA supplemented with different NaCl concentrations (3% and 10%) and adjusted to pH 4, 7, and 9. Temperature tolerance was evaluated by incubating the strain on MA at temperatures ranging from 25°c to 60°c at 5°c intervals (Yang et al., 2025).
For molecular identification, isolates obtained from the sixteen marine environmental and fermented seafood samples were subjected to 16S ribosomal RNA (16S rRNA) gene sequencing. Single colonies of each isolate were submitted to Macrogen Inc. (Seoul, South Korea) for PCR amplification and sequencing of the 16S rRNA gene. The 16S rRNA gene was amplified using the universal bacterial primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-CGGTTACCT TGTTACGACTT-3′). The resulting sequences were compared against publicly available 16S rRNA gene sequences in the EzBioCloud database (http://www.ezbiocloud.net) to determine taxonomic identity based on sequence similarity. A circular phylogenetic tree was constructed using the Neighbor-Joining (NJ) method (Saitou & Nei, 1987). Evolutionary distances were calculated using the Maximum Composite Likelihood method (Tamura et al., 2004). All evolutionary analyses were performed using MEGA version 12 (Kumar et al., 2024).
The production of extracellular hydrolytic enzymes (protease, amylase and lipase) by the isolated halophilic microorganisms was evaluated using solid media containing specific substrates for each enzyme. For the detection of protease activity, MA was supplemented with 20% (w/v) skim milk (BD, USA) as the substrate. Amylase activity was assessed on MA containing 0.2% (w/v) soluble starch (BD, USA), and lipase activity was evaluated using MA plates supplemented with 1% (v/v) Tween 80 (BD, USA). Each isolate was spot-inoculated onto the respective plates and incubated at 37°c for 7 days under aerobic conditions. After incubation, the diameter of the clear zone surrounding each colony was measured to determine enzyme activity. Hydrolytic activity was semi-quantitatively graded based on clear zone as follows: +++ (strong activity, >7 mm), ++ (moderate activity, 4–6 mm), and + (weak activity, 1–3 mm) (Yang et al., 2025).
The ability of the isolated strains to produce auxin (indole-3-acetic acid; IAA) was evaluated using a colorimetric assay. Each pure colony was inoculated into Marine Broth (MB; BD Difco, USA) supplemented with 0.1% (w/v) L-tryptophan (Sigma-Aldrich, USA) and incubated at 37°c for 5 days under aerobic conditions. After incubation, cultures were centrifuged at 10,000×g for 10 min to obtain the cell-free supernatant. For IAA detection, 400 μL of each culture supernatant was mixed with 800 μL of Salkowski reagent (prepared by combining 50 mL of 35% HClO4 and 1 mL of 0.5 M FeCl3). The mixtures were incubated in the dark at room temperature for 30 min to allow color development, after which absorbance was measured at 535 nm using a spectrophotometer. IAA production was semi-quantitatively classified based on the absorbance value as follows: +++ (strong production, >1.0), ++ (moderate, 0.5–1.0), and + (weak, 0.3–0.5) (Yang et al., 2025).
Results and Discussion
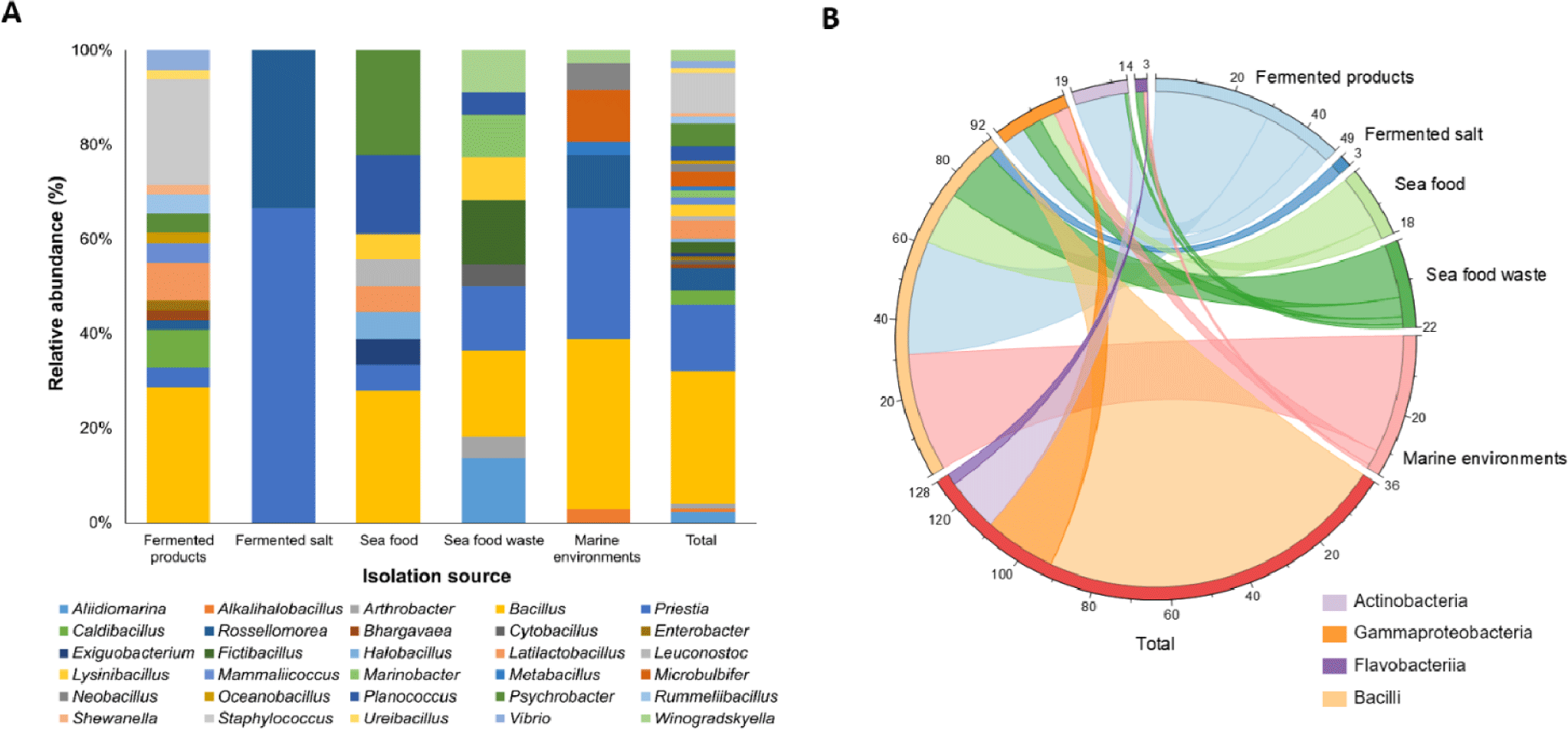
A total of sixteen samples, including seawater, tidal flat sediments, and various fermented seafoods, were collected from different sites in Sacheon City to isolate aerobic halophilic microorganisms with potential application in the food industry. Each sample was serially diluted and spread on MA, and colonies exhibiting distinct morphological characteristics (color, size, and texture) were selected. Pure cultures were obtained through repeated single-colony streaking on fresh MA plates. As shown in Fig. 1 and Supplementary Table 1, 36 strains were isolated from marine environmental samples, and 92 strains were obtained from fermented seafood samples, yielding a total of 128 halophilic isolates specific to the Sacheon region.
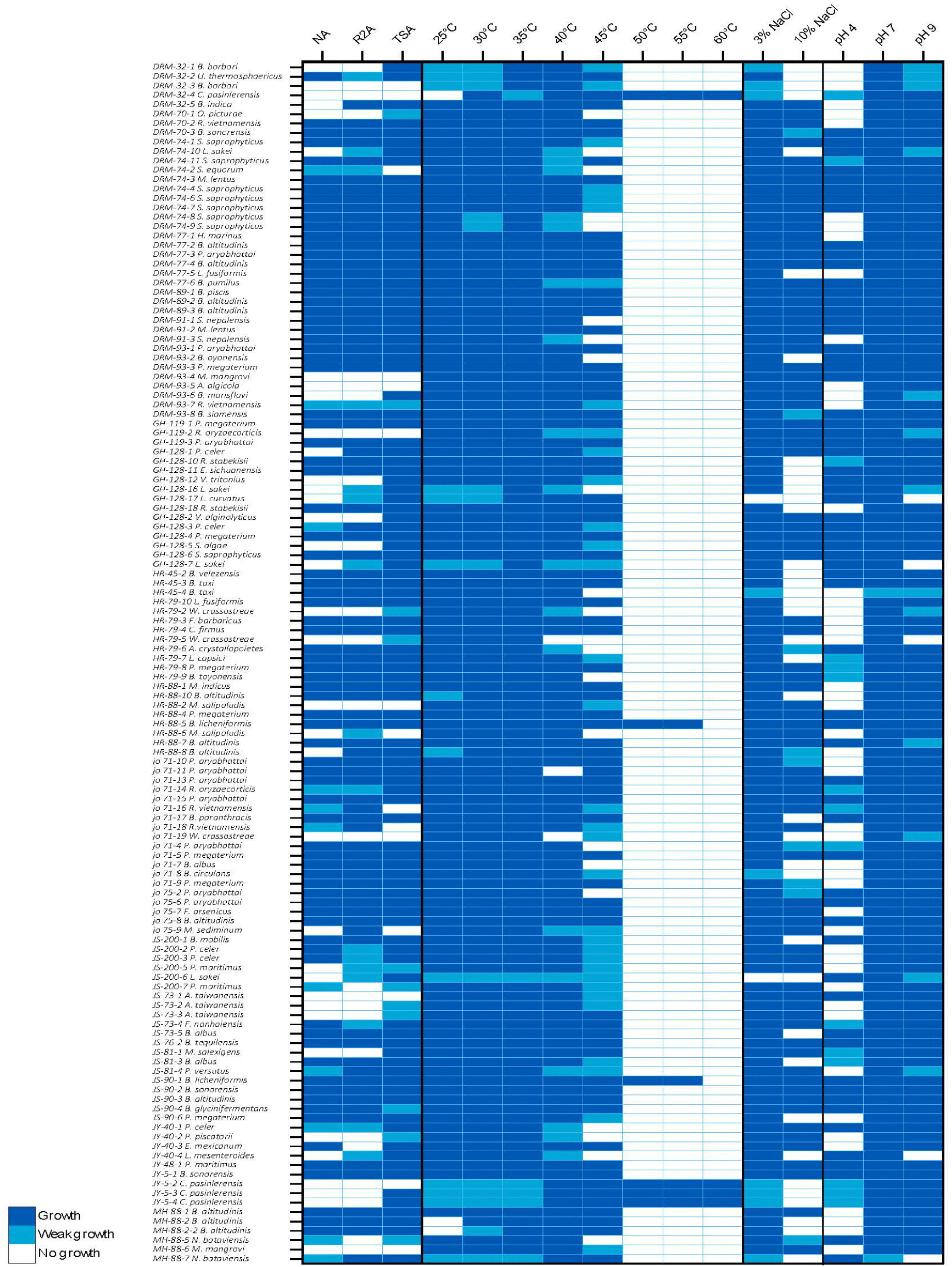
Because marine agar, composed primarily of inorganic salts, is well-suited for culturing marine halophiles, the growth ability of the isolates was further examined on complex media (NA, R2A, and TSA) commonly used for industrial-scale cultivation. Among the 128 isolates, 118 strains (including weak growth) were able to grow on at least one of these three media, indicating that medium composition affects the isolation efficiency of halophiles to some extent (Fig. 2). Based on these results, MA was confirmed to be the most suitable isolation medium for halophilic bacteria from marine and fermented seafood samples. To determine pH adaptability, isolates were cultivated on MA plates adjusted to pH 4, 7, and 9. All isolates exhibited growth (including weak growth) at pH 7, whereas 79 strains grew at pH 4 and 123 strains grew at pH 9. Notably, 75 isolates demonstrated tolerance across all three pH conditions (Fig. 2). Temperature tolerance tests revealed that all isolates grew optimally at 30–35°c, and 106 strains displayed growth at 45°c. Interestingly, six isolates grew at 50–55°c, and four strains of Caldibacillus pasinlerensis were capable of growth even at 60°c (Fig. 2). Salt tolerance tests showed that 126 isolates could grow in media containing >3% NaCl, and 90 isolates exhibited growth at NaCl concentrations exceeding 10% (Fig. 2).
Although numerous studies have reported the isolation and identification of microorganisms from individual fermented seafoods such as anchovy sauce (myeolchi-aekjeot), sand lance (bandengi), oyster viscera, hairtail viscera, and clam (bajirak) products, comparative analyses of microbial distribution and characteristics among multiple fermented seafoods collected from a specific region remain limited. Likewise, studies isolating microorganisms from oyster shells, crushed shell debris, or tidal mud from small islands are also scarce. In recent years, however, regional initiatives in areas such as Sunchang, Jeju, and Sacheon have emphasized the exploration of region-specific microorganisms and their potential industrial applications. In Sunchang, key microbial strains involved in traditional fermented condiments (gochujang, doenjang, and ganjang) have been systematically characterized, while researchers have investigated the influence of unique environmental factors (volcanic soil, marine ecosystems) on microbial diversity in Jeju and published a three-volume Jeju Native Microorganism Atlas (Yang et al., 2025). In addition, microbial community analyses of various jeotgal types, including anchovy, sea squirt and shrimp fermentations (Song et al., 2018), studies on microbial profiling in oyster and clam samples (Kim et al., 2024), and reports of halophilic isolates from diverse marine environments such as sediments, seawater, shells, and tidal flats (Lee et al., 2023) have provided valuable foundations for expanding regional microbial resource studies. Following this trend, the Sacheon Microbial Fermentation Foundation has focused not only on the isolation and identification of microorganisms from regional fermented foods but also on the characterization of their industrially valuable traits. In this study, 128 halophilic bacterial strains exhibiting diverse media adaptability and strong extracellular enzymatic activities were secured from the marine and fermented seafood resources of Sacheon. These results provide a meaningful foundation for the industrial utilization of region-specific microbial resources.
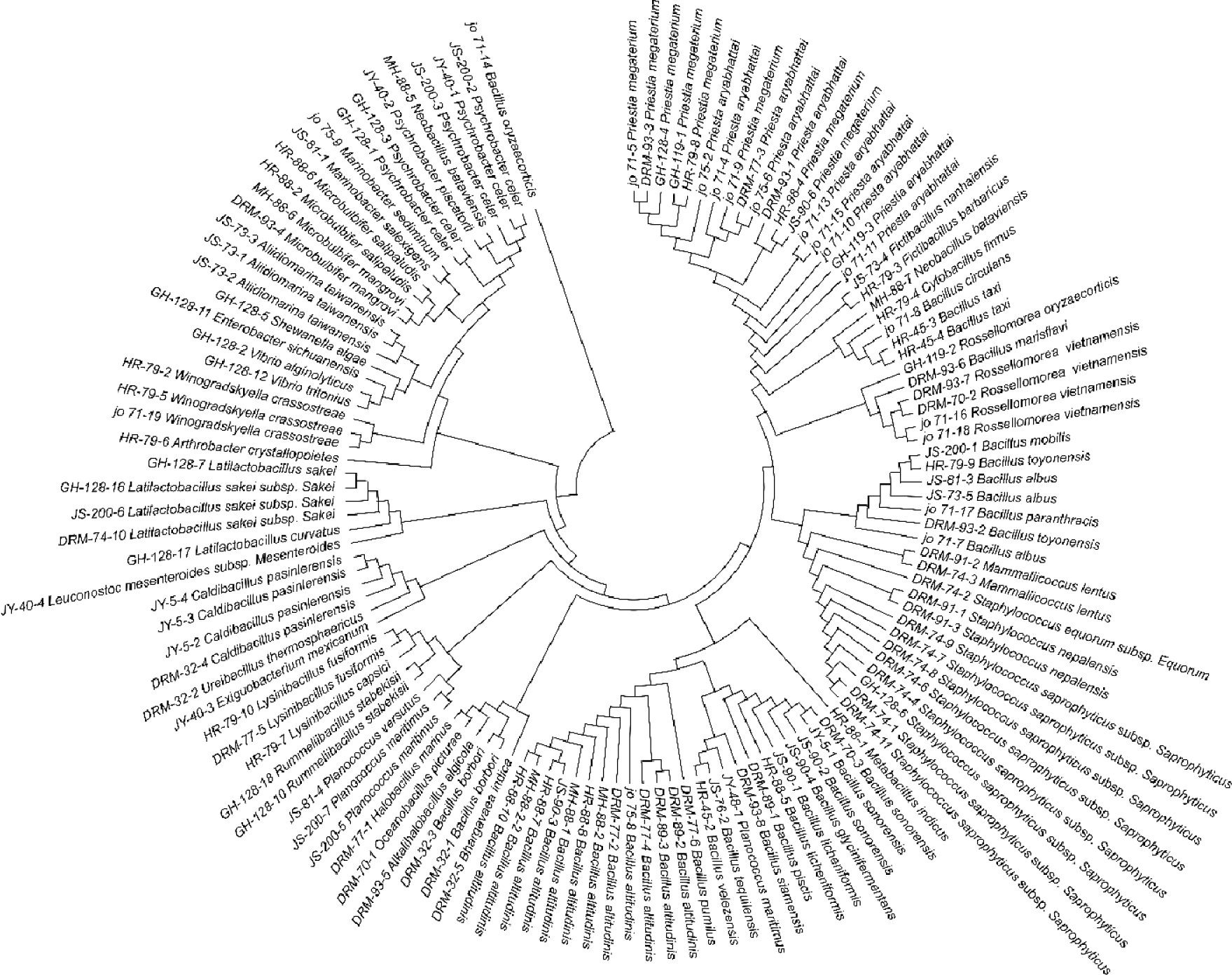
A total of 128 aerobic bacterial strains isolated from marine environmental samples and fermented seafood products collected in Sacheon City were identified using the EzBioCloud database based on 16S rRNA gene sequences. The isolates were classified into four phyla, 15 families, 30 genera, and 61 species (Fig. 1). Detailed information regarding each isolate, its phylogenetically closest type strains, and sequence similarity values is provided in Supplementary Table 1. To elucidate evolutionary relationships among the isolates, a phylogenetic tree was constructed based on the 16S rRNA gene (Fig. 3). The phylum Bacillota was dominant, accounting for 82.0% of all isolates. Within Bacillota, the family Bacillaceae was the most abundant (76.2%), followed by Staphylococcaceae (12.4%), Lactobacillaceae (4.8%), Planococcaceae (3.8%), with minor contributions (~1% each) from Caryophanaceae, Exiguobacteraceae, and Leuconostocaceae. In total, 48 species across 21 genera belonging to seven families were identified.
Among the Bacillaceae family, which exhibited the highest isolation frequency, the genus Bacillus is widely known to be prevalent in marine and freshwater organisms and is frequently isolated from various traditional fermented foods, particularly those derived from marine environments, fermented agricultural materials, and jeotgal (salted seafood). Therefore, the predominance of Bacillaceae in the marine and fermented samples from Sacheon, a region characterized by a complex ria-type coastline and diverse coastal habitats, is well supported by previous findings (Yoon et al., 2001; Dharaneedharan & Heo, 2016). In this study, halophilic bacteria belonging to the Bacillaceae family were abundantly isolated from two marine environmental samples and 13 fermented seafood samples (including various types of jeotgal and oyster shell samples) among the 16 total sampling sites in Sacheon. Their predominance likely reflects the high-salinity conditions characteristic of fermented seafood environments, which favor the growth of Bacillus species known for robust pH regulation and antimicrobial potential (Kim et al., 2010; Kim et al., 2013).
Among the genus Bacillus, Bacillus altitudinis was identified in 12 isolates (33.3%) derived from five different marine and fermented seafood samples collected in the Sacheon region. As reported by Yang et al. (2025), B. altitudinis is considered a representative strain well adapted to and stable in the long-term fermentation environments of Sacheon, including both fermented agricultural and seafood products. Importantly, several isolates obtained in this study belonged to taxa designated as Generally Recognized As Safe (GRAS) or Qualified Presumption of Safety (QPS) by the European Food Safety Authority (EFSA BIOHAZ Panel, 2025), underscoring their potential safety and suitability for industrial use. Representative species include Bacillus licheniformis,Bacillus sonorensis,Bacillus toyonensis,Latilactobacillus curvatus,Latilactobacillus sakei, and Leuconostoc mesenteroides. Their presence highlights the value of Sacheon-derived microbial resources for applications in food fermentation, bioprocessing, and other biotechnology sectors.
Marine environments and fermented seafood products are characterized by high salinity, mineral-rich, and enhanced umami flavor, with fermentation processes accelerated by coastal climatic conditions and influenced by the diverse enzymatic activities of associated microorganisms (Park et al., 2025). To evaluate the potential industrial applicability of the halophilic bacterial isolates from Sacheon as potential producers of hydrolytic enzymes for the food industry and as biofertilizer candidates for agricultural applications, their extracellular enzyme activities and auxin (IAA) production abilities were analyzed. Among the 128 isolates, 86 strains (67.2%) exhibited at least one type of hydrolytic enzyme activity (Supplementary Table 1). Specifically, 69 strains showed protease activity, 49 strains exhibited amylase activity, and 14 strains displayed lipase activity. Notably, 42 isolates exhibited two or more enzymatic activities. The most common combination was concurrent amylase and protease activity, observed in 21 isolates (Fig. 4a). Four strains (Bacillus tequilensis JS-76-2, Microbulbifer mangrovi DRM-93-4, MH-88-6, and Planococcus maritimus JY-48-1) exhibited all three hydrolytic activities, indicating broad metabolic versatility (Supplementary Table 1). In particular, B. tequilensis has previously been reported as a representative halophilic strain well adapted to long-term fermentation environments in the Sacheon region and is known for robust enzymatic activity and high environmental adaptability (Kim et al., 2010; Kim et al., 2013). Therefore, B. tequilensis and other halophilic enzyme-producing isolates identified in this study are considered valuable microbial resources for biotechnological processes and food fermentation industries. When compared with previous reports, the proportion of strains exhibiting hydrolytic enzyme activity in this study (67.2%) is consistent with values typically reported for halophilic microorganisms in high-salinity habitats, although it is lower than the 92.2% recently documented for isolates from fermented agricultural products in the Sacheon region (Yang et al., 2025). Reported proportions from other salt-associated environments include 77.2% in strains from solar salt fields in Shinan and Gomso (Lee et al., 2020a), 77.6% in isolates from Jeju coastal ranch areas (Lee et al., 2020b), 64.0% in strains from South Sea and Tsushima seafoods (Jeong et al., 2021), and 67.4% in isolates from southwestern coastal soils of Jeju (Lee et al., 2022). These variations are likely attributable to differences in sample substrates and physicochemical conditions, as our study encompassed marine environmental and fermented seafood samples, whereas Yang et al. (2025) focused on carbohydrate-rich agricultural fermentations that selectively enrich strong protease- and amylase-producing taxa.
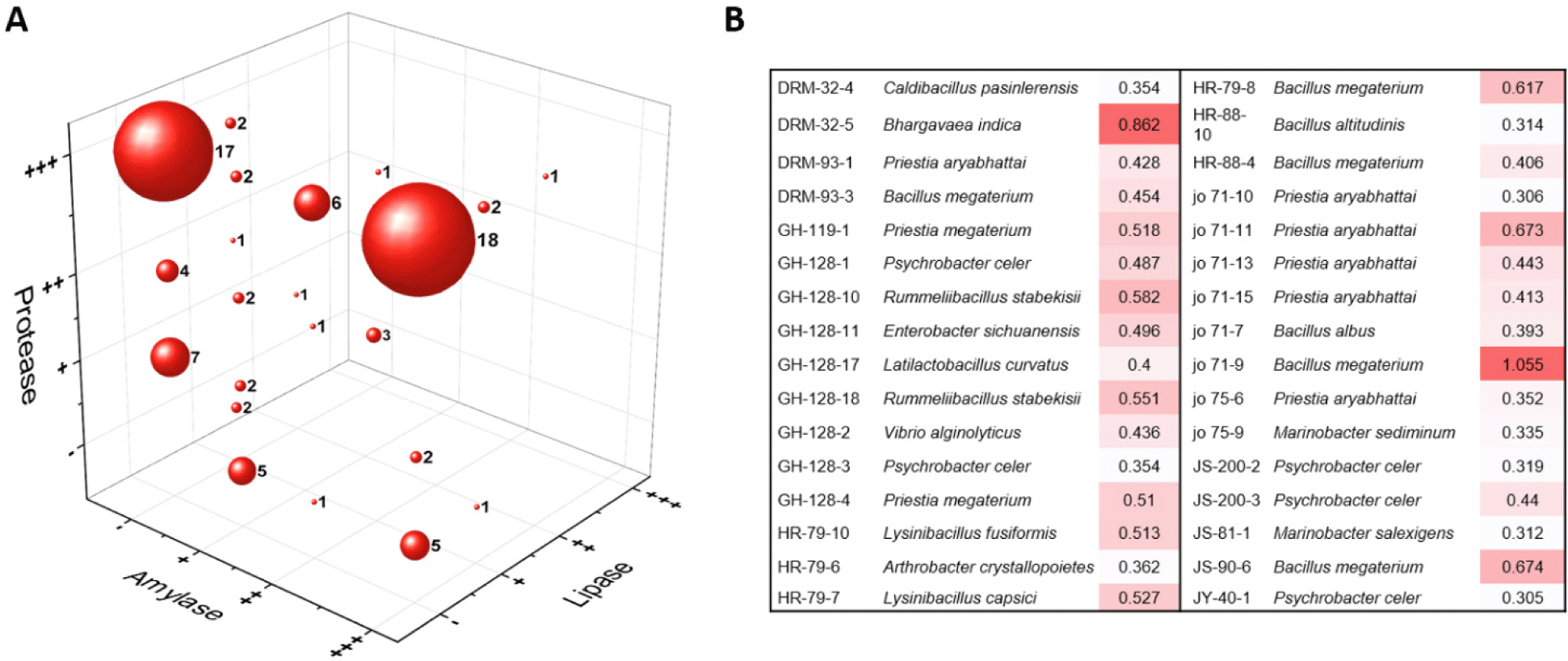
IAA, the primary form of microbial auxin, plays essential roles in plant growth and development, including cell elongation, root initiation, seed germination, fruit enlargement, photosynthetic enhancement, stress tolerance, and nitrogen fixation (Woo & Kim, 2007). A total of 32 isolates demonstrated IAA production capabilities (Fig. 4b). The highest IAA production activity was observed in Bhargavaea indica strain DRM-32-5, isolated from anchovy jeotgal, and Priestia megaterium (formerly Bacillus megaterium) strain JO71-9, isolated from near the Bito Coastal Breakwater. These findings highlight the potential of isolated strains as microbial biofertilizers, particularly for coastal agriculture and saline soils where conventional plant growth–promoting microorganisms have limited survival (Etesami, 2020). In addition to their agricultural relevance, the inherent stress-tolerance of these halophiles positions them as promising candidates for environmental biotechnology, including the treatment of saline wastewater and the biodegradation of seafood-processing effluents (Sadeghi et al., 2019; Mainka et al., 2021). Halophilic bacteria are also known to harbor distinctive stress-adaptive biosynthetic pathways, offering valuable opportunities for genome mining, enzyme engineering, and metabolic pathway analysis aimed at discovering novel salt-stable enzymes and bioactive metabolites (DasSarma & DasSarma, 2015; Dutta et al., 2022).
The findings of this study contribute to expanding the diversity of indigenous Korean microbial resources derived from the marine environments and fermented seafoods of the Sacheon region. Collectively, the isolates secured in this study represent an important microbial resource with substantial potential for the development of industrial biocatalysts, environmentally friendly microbial formulations, and biofertilizer applications. All strains isolated in this study have been deposited in the Microbial Resource Division of the Korea Research Institute of Bioscience and Biotechnology (KRIBB) under the Microbial Value Enhancement Program (Supplementary Table 1), ensuring their long-term accessibility as region-specific biological resources.







