Introduction
In 2002, a working group of international scientists from the Food and Agriculture Organization of the United Nations (FAO) and the World Health Organization (WHO) discussed the field of probiotics and developed guidelines to aid interpretation, defining probiotics as “live microorganisms which when administered in adequate amounts confer a health benefit on the host” (FAO/WHO, 2002). Lactic acid bacteria are prevalent in probiotics because of their historical use and convenience (Żukiewicz-Sobczak et al., 2014). These organisms are naturally found in the human gastrointestinal tract and have been utilized in various dairy industry processes, which have established methods for their large-scale manufacturing and preservation (Hove et al., 1999; Colombo et al., 2018).
While probiotics have been considered safe and recognized as generally recognized as safe strains, their safety has been recently scrutinized due to significant advancements in the field (Sanders et al., 2010). As a result, the FAO/WHO guidelines recommend utilizing clinically standardized techniques to assess the safety of probiotics (Hill et al., 2014). To ensure the safety of probiotics for consumption by humans and animals, guidelines recommend determining the antibiotic resistance patterns of probiotic strains and ensuring that there are no acquired, or transferable resistance factors present (Rychen et al., 2018). This is important to prevent the spread of antibiotic resistance and ensure that probiotics do not contribute to the development of antibiotic-resistant bacteria (Imperial & Ibana, 2016). It is also important to follow good manufacturing practices and quality control procedures to ensure the safety and efficacy of probiotic products (Sanders et al., 2010).
Lactiplantibacillus plantarum and Levilactobacillus brevis, currently widely used probiotics, were reclassified as new L. plantarum and L. brevis, distinct from other Lactobacillus species, based on whole genome sequencing (de Vries et al., 2006; Zheng et al., 2020). Many studies have demonstrated the safety of L. plantarum and L. brevis (Kwon et al., 2021; Lee et al., 2025). However, concerns remain regarding the inherent risks of probiotics, including toxicity, biofilm formation, and the potential risks of antibiotic resistance genes through mutations or horizontal gene transfer between pathogens (Merenstein et al., 2023; Haranahalli Nataraj et al, 2024; Xu et al., 2025). Therefore, safety assessment of probiotics is essential in various industries.
In a previous study, L. plantarum KU15149 isolated from diced-radish kimchi exhibited antioxidant and anti-inflammatory effects, and did not produce β-glucuronidase, a carcinogenic enzyme, nor show antibiotic resistance (Han et al., 2020). In addition, L. brevis KU15176 was isolated from cabbage kimchi, and the heat-killed strain was confirmed to exert an anticancer effect by inducing the expression of apoptosis-related genes and increased caspase activity in a human gastric adenocarcinoma cancer cell line (Hwang et al., 2022). Along with the previous study, the safety of L. plantarum KU15149 and L. brevis KU15176 as potential probiotic strains was evaluated. The assessment involved examining their phenotype, which included antibiotic resistance, hemolysis, and production of toxic metabolites, as well as genotype, which involved checking for the presence of virulence and antibiotic resistance genes. FAO/WHO guidelines were followed during the evaluation process.
Materials and Methods
L. plantarum KU15149 and L. brevis KU15176 were provided by Konkuk University (Seoul, Korea) and were cultivated in MRS medium (Oxoid, Basingstoke, Hampshire, UK) at 37°c for 24 h. The positive control strain for each assay, Lactococcus lactis subsp. lactis CAB 1001 was grown in MRS broth at 37°c for 24 h. Brevibacillus parabrevis KCCM 42421, Streptococcus pyogenes KCCM 11873, and Escherichia coli ATCC 10536 were grown in tryptic soy broth (TSB, BD Biosciences, Franklin Lakes, NJ, USA) at 30°c for 24 h. Proteus vulgaris KCCM 40221 and Klebsiella pneumoniae subsp. pneumoniae KCCM 60022 were grown in nutrient broth (BD, USA) at 30°c for 24 h and were purchased from the Korean Culture Center of Microorganisms (KCCM, Seoul, Korea).
Hemolysis occurs due to the hemolysin produced by bacteria and it degrades red blood cells in the human body. In terms of food safety, hemolytic activity must be tested. The assay was performed by inoculating bacterial cells inoculated on blood agar plates supplemented with 5% (v/v) sheep blood and incubated anaerobically at 37°c for 48 h. Hemolysis was detected by the clear zones around the colony.
Mucin degradation was performed by inoculating 1% (v/v) overnight grown culture medium into four different media (basal MRS, basal MRS including 0.3% mucin, basal MRS including 1% glucose, and basal MRS including 0.3% mucin and 1% glucose) and incubating at 37°c for 8 h and 24 h. To assess bacterial mucin degradation activity of bacteria, absorbance was measured at 600 nm (Shimadzu Co., Kyoto, Japan), and the pH of the culture medium was measured (Mettler-Toledo Inc., Columbus, OH, USA).
Gelatin liquefaction was conducted by streaking bacterial cells on MRS slants containing 12% gelatin and incubating at 37°c for 72 h. B. parabrevis KCCM 41421, as a positive control for the gelatin liquefaction test, was incubated in TSB at 37°c for 72 h. Gelatin liquefaction ability was observed after 30 min of incubation on ice.
Urease activity was assessed on Christensen’s urea agar (0.1 g peptone, 0.1 g dextrose, 0.5 g sodium chloride, 0.2 g KH2PO4, 2 g urea, 0.0012 g phenol red, 1.5 g agar, per 100 mL). L. plantarum KU15149, L. brevis KU15176, and P. vulgaris KCCM 40221 (positive control) were inoculated and incubated at 37°c for 24 h. Urease activity was determined by the color change of the medium.
Indole production was determined by inoculating bacteria and E. coli ATCC 10536 (positive control) into tryptophan medium (10 g casein enzyme hydrolysate, 5 g NaCl, 1 g DL-tryptophan, per liter) and incubating at 37°c for 18 h. After incubation, five drops of Kovac’s reagent (a mixture of 10 g p-dimethylaminobenzaldehyde, 150 mL butanol, and 50 mL hydrochloric acid) were added, and the development of a red color was considered a positive control.
Enzymatic activities were assessed using an API-ZYM kit (BioMerieux, Marcy-L’Etoile, France) according to the manufacture’s instruction. Overnight grown bacterial cultures were diluted with 2 mL of sterile saline to adjust the turbidity to McFarland 5-6. Afterwards, 65 μL of the suspension was dispensed into each API strip and incubated at 37°c for 4 h. One drop each of ZYM A and ZYM B reagents was then added, and the color change of each well was examined after 5 min.
To determine bile salt deconjugation, L. plantarum KU15149 and L. brevis KU15176 were streaked onto MRS agar plates supplemented with 0.5% taurodeoxycholic acid (bile acid, Sigma-Aldrich, St. Louis, MO, USA) and incubated anaerobically at 37°c for 48 h. Bile salt deconjugation was indicated by the precipitation around the colonies.
Antibiotic susceptibility of L. plantarum KU15149 and L. brevis KU15176 was determined using E-test strip (BioMerieux). Each colony of L. plantarum KU15149 and L. brevis KU15176 was inoculated into MRS broth and incubated at 37°c for 20 h. A 100 μL aliquot of the culture diluted in sterile 0.85% (w/v) NaCl to match the turbidity of a 3.0 McFarland standard was spread onto MRS agar plates. Each E-test strip was then placed in the center of the plate and incubated at 37°c for 72 h. The number at the end of the strip within the inhibition zone was defined as the minimum inhibitory concentration (MIC). The antibiotics used in this assay were ampicillin, vancomycin, gentamycin, kanamycin, streptomycin, erythromycin, clindamycin, tetracycline, and chloramphenicol, in accordance with the European Food Safety Authority (EFSA) guidelines (Additives & Feed, 2012).
Genomic DNA of L. plantarum KU15149 and L. brevis KU15176 was extracted using the AccuPrep® Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea) according to the manufacturer’s instructions. The extracted DNA was used to identify antibiotic resistance genes with My Cycler™ (Bio-Rad, Hercules, CA, USA). Primer sequences and target sizes for each antibiotic resistance genes are shown in Table 3.
Whole-genome sequencing of L. plantarum KU15149 and L. brevis KU15176 was performed by CJ Bioscience Inc. (Seoul, Korea) using lllumina MiSeq platform. The genomic sequences obtained were analyzed for antibiotic resistance and virulence genes using the ResFinder v.4.1 (https://cge.cbs.dtu.dk/services/ResFinder/) (Zankari et al., 2012), the Comprehensive Antibiotic Resistance Database (CARD) (https://card.mcmaster.ca/) (Alcock et al., 2023), and the Resistance Gene Identifier (RGI) v.5.2.0 (Alcock et al., 2023). Default settings were used for the ResFinder analysis, wherease perfect, strict, and loose hits were applied for the RGI analysis. Virulence genes were identified using VirulenceFinder 2.0 (http://cge.cbs.dtu.dk/services/VirulenceFinder/) by screening for homologous sequences to virulence genes of E. coli, Listeria spp., Staphylococcus aureus, and Enterococcus spp. The analytical database system was provided by the Center for Genomic Epidemiology. Thresholds of a minimum percent identity (%ID) of 90% and a minimum sequence length of 60% were applied for identifying homologous sequences.
The quantification of D-/L-lactic acid was performed using an assay kit (Roche Diagnostics International Ltd, Rotkreuz, Switzerland) according to the manufacturer’s instructions. Absorbance was measured three times at 340 nm using a spectrophotometer (Bio-Tek Instruments, Winooski, VT, USA). The first mixture solution consisted of 1 mL of solution 1, 200 μL of solution 2, 20 μL of solution 3, and 100 μL of culture supernatant with 900 μL of d-H2O. The first sample was measured after 5 min (A1). The second sample was measured after 30 min after adding 20 μL of solution 4 (A2). The last sample was measured after 30 min after adding 20 μL of solution 5 (A3). Production of D-/L-lactate was calculated in mM according to the manufacturer’s instructions.
The biogenic amines assessed in this study were putrescine, β- phenylethylamine, spermidine, serotonin, histamine, tyramine, tryptamine, and agmatine (Sigma-Aldrich). The stock solution of biogenic amines was adjusted to 1,000 ppm with 0.1 N HCl. One percent of overnight grown bacteria was inoculated into fresh MRS broth consisting of 200 ppm of arginine, ornithine, histidine, tyrosine, tryptophan, lysine, and phenylalanine, which are the precursors of biogenic amines assessed in this assay. The cultures were incubated at 37°c for 20 h. For derivatization at 45°c for 1 h, 1 mL of culture medium, 500 μL of saturated sodium carbonate solution, and 800 μL of 1% (w/v) dansyl chloride acetone solution was mixed. After derivatization, 500 μL of 10% (w/v) proline and 5 mL of diethyl ether were added and mixed by vortexing for 10 min. The upper layer of the mixture was transferred to a new tube and dried with nitrogen gas. One milliliter of acetonitrile (ACN, Sigma-Aldrich) was then added to dissolve the dried sample. The dissolved solution was filtered using 0.22 μm SmartPor®-II PVDF Syringe filter (Woongki Science Co., Ltd., Seoul, Korea). The filtered solution was analyzed by high-performance liquid chromatography (HPLC) with a Dionex UltiMate 3000 variable wavelength detector (Thermo Fisher Scientific, Waltham, MA, USA). Detection of biogenic amine was performed using 250×4.6 mm, 5 μm, Agilent 5 TC-C18 column (Agilent Technologies, Santa Clara, CA, USA), with UV detection at 254 nm. HPLC analysis was carried out using a gradient elution, with the mobile phase consisting of acetonitrile and water. The total running time was 30 min, and the mobile phase gradient was as follows: 0–2 min, 55% ACN/45% water; 2–5 min, 65% ACN/35% water; 5–10 min, 65% ACN/35% water; 10–15 min, 80% ACN/20% water; 15–20 min, 80% ACN/20% water; 20–25 min, 55% ACN/45% water; 25–30 min, 55% ACN/45% water. The flow rate was 1 mL/min, and the column temperature was 40°c.
Caco-2 cells (ATCC® HTB-37™), a human colon adeonocarcinoma cell line, were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in Dulbecco’s modified Eagle’s medium (Gibco, Grand Island, NY, USA) supplemented with 10% (v/v) of fetal bovine serum (Corning Inc., Corning, NY, USA) and 1% (v/v) penicillin-streptomycin (Gibco) in an incubator with 5% CO2 at 37°c (Thermo Fisher Scientific). 0.25% Trypsin-EDTA (Gibco) was used for cell detachment.
For the cytotoxicity assay, K. pneumoniae subsp. pneumoniae KCCM 60022 was used as a pathogenic strain and cultured in NB medium at 37°c for 16 h. Caco-2 cells were seeded into 96-well plates at a density of 5×104 cells per well and incubated in a 5% CO2 incubator at 37°c for 20 h. L. plantarum KU15149 and L. brevis KU15176 were cultured overnight, then incubated in fresh MRS broth until reaching a density of 108 CFU/mL. Afterwards, 5 mL of bacterial culture was collected by centrifugation at 22,673×g for 1 min at 4°c and washed three times with Dulbecco’s phosphate- buffered saline (DPBS, Welgene, Daegu, Korea). In each well, bacteria were added at an MOI of 250 (multiplicity of infection, bacterial cells/Caco-2 cells) and incubated at 37°c for 24 h. After incubation, the cells were washed three times with DPBS and then incubated with 20 μL of EZ-CYTOX (DoGenBio, Seoul, Korea) at 37°c for 30 min. Absorbance was measured at 450 nm using an epoch microplate spectrophotometer (Bio-Tek Instruments).
All experiments were performed in triplicate, and the results are expressed as the mean±standard deviation. Statistical significance was evaluated by one-way or two-way analysis of variance (ANOVA) using SPSS version 25 (IBM Co., Armonk, NY, USA) and GraphPad Prism version 10.3.1 (GraphPad Software, Boston, MA, USA). Post hoc analyses, including Dunnett’s, Duncan’s, and Sidak’s multiple comparison tests, were applied to determine significant differences among groups.
Results and Discussion
To test the safety of L. plantarum KU15149 and L. brevis KU15176, hemolytic activity, mucin degradation, gelatin liquefaction, urease activity, indole production, β-glucuronidase activity, bile salt deconjugation, and enzymatic activities were determined (data not shown).
In the hemolytic activity assay, S. pyogenes KCCM 11873 served as the positive control. Neither L. plantarum KU15149 nor L. brevis KU15176 exhibited hemolytic activity, whereas the positive control demonstrated clear hemolysis. The probiotic strains of Enterococcus faecium RM11 and Limosilactobacillus fermentum RM28 showed γ- hemolysis (Thirabunyanon et al., 2009). Jehma et al. (2025) reported that L. plantarum LP8 displayed α-hemolysis activity; however, no virulence genes were detected in its genome sequence.
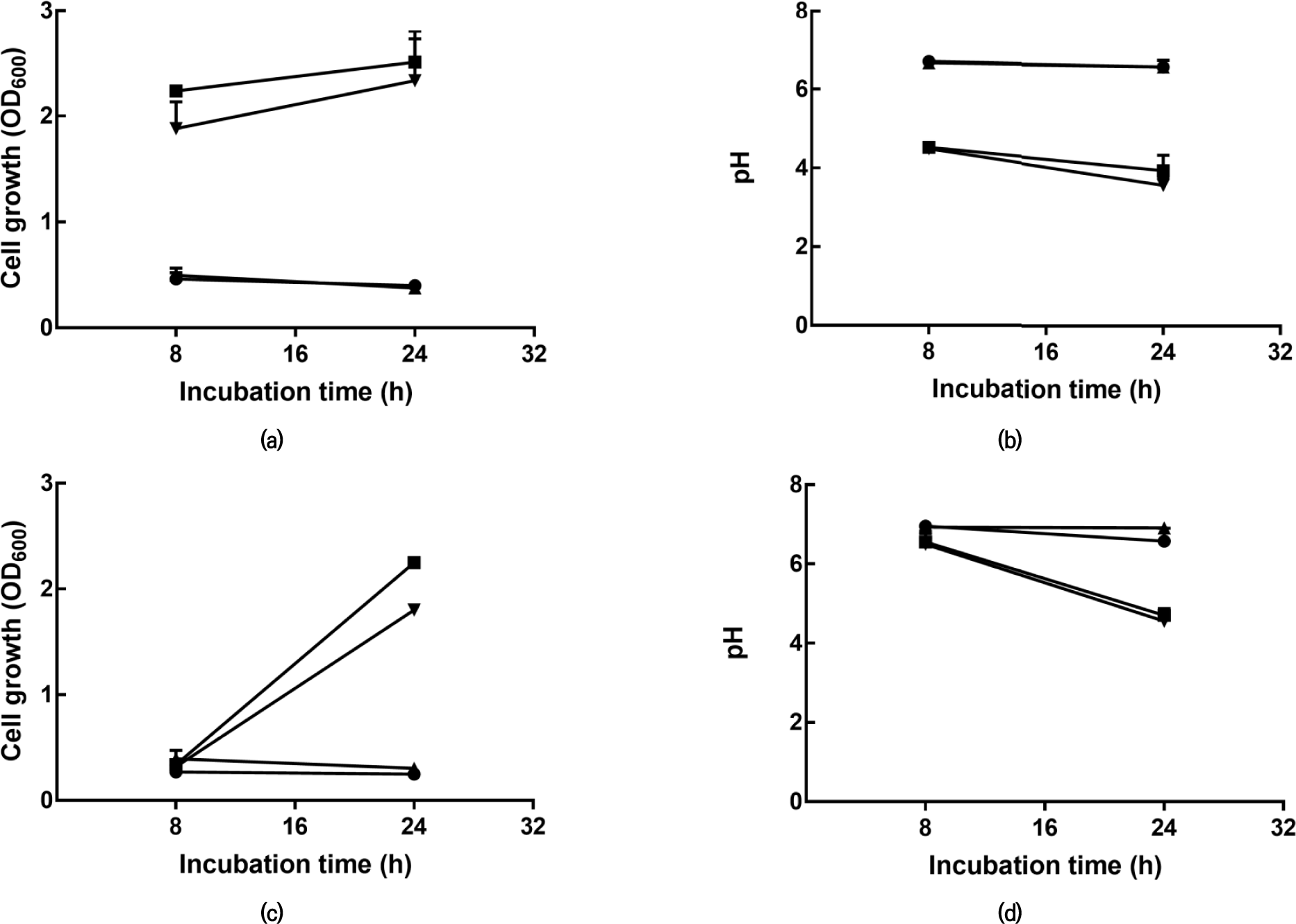
To determine mucin degradation activity, bacterial growth was examined after incubation in four different media. Growth was assessed by measuring absorbance at 600 nm and pH. As a result, based on OD600 and pH values after 8 h and 24 h of incubation, the growth of L. plantarum KU15149 and L. brevis KU15176 in basal MRS containing 0.3% mucin was insignificant compared to that in the medium containing glucose (Fig. 1). It has not been confirmed that lactic acid bacteria directly degrade mucin.
In bacteria with gelatinase activity, a phenotypic change occurs as the medium liquefies rather than remaining solid, as observed with P. vulgaris KCCM 40221. However, in media inoculated with L. plantarum KU15149 and L. brevis KU15176, the medium remained solid after incubation, indicating that gelatin liquefaction did not occur.
Urease activity was determined by assessing whether bacteria inoculated in the medium released urease to decompose urea among the medium components. The indicator phenol red appears yellow at pH 6.8 or lower and red at pH 8.4 or higher. No color change was observed in L. plantarum KU15149 and L. brevis KU15176, indicating that urease activity was absent.
Tryptophanase converts tryptophan into indole, pyruvic acid, and ammonia (Watanabe & Snell, 1972). Indole reacts with p-dimethylaminobenzaldehyde in Kovac’s reagent to form a red quinone-based compound in the upper layer of the medium. The result showed that no color change was found in L. plantarum KU15149 and L. brevis KU15176 whereas E. coli ATCC 10536, positive strain in this assay, caused a color change to pink.
Bile salts emulsify fats and facilitate lipase activity in fatty acid decomposition. Primary bile salts, such as cholic acid and chenodoxyholic acid, are convert to secondary bile salts by 7α- dehydroxylase (Kang et al., 2019). Secondary bile salts, including deoxycholic acid and lithocholic acid, are associated with an increased risk of gastrointestinal cancer (Domellöf et al., 1980). Therefore, excessive intake should be avoided. In a previous study (Lee et al., 2025), colonies were observed around Lactococcus lactis subsp. lactis CAB 1001, which was used as a positive control for the bile salt deconjugation test. Neither of L. plantarum KU15149 nor L. brevis KU15176 has bile salt hydrolase activity.
The activity of β-glucuronidase in the gut can lead to the production of toxic metabolites from compounds such as estrogen, which have been implicated in colorectal cancer development (Kim & Jin, 2001). Enzymatic activities were assessed using API-ZYM kit, and L. plantarum KU15149 and L. brevis KU15176 tested negative for β-glucuronidase, indicating that they do not produce toxic metabolites (Table 1).
Both L. plantarum KU15149 and L. brevis KU15176 were susceptible to nine antibiotics, including ampicillin, gentamycin, kanamycin, streptomycin, erythromycin, clindamycin, tetracycline, and chloramphenicol (Table 2). Vancomycin was not required for both strains and streptomycin were not required for L. plantarum KU15149. As a result, the MIC of L. plantarum KU15149 and L. brevis KU15176 was lower than the cut-off values established in EFSA guideline, except for vancomycin, whereas L. plantarum KU15149 was found to be resistant to vancomycin. Previous study elaborates that due to its intrinsic resistance to glycopeptides, L. plantarum could show MIC to vancomycin (Perichon & Couvalin, 2000).
To verify any antibiotic resistance genes (ARG), polymerase chain reaction (PCR) was performed to analyze resistance genes including ampicillin, chloramphenicol, clindamycin, erythromycin, gentamycin, kanamycin, streptomycin, tetracycline, and vancomycin. As a result of the experiment, all antibiotic resistance genes except the vancomycin resistance gene were not detected by PCR in L. plantarum KU15149 and L. brevis KU15176, ant the results are summarized in Table 3. VanX showed positive in L. plantarum KU15149 but as vancomycin is not a required antibiotic in lactic acid bacteria, no ARGs were found in L. plantarum KU15149 and L. brevis KU15176. As every antibiotic tests showed that none of antibiotics resistance genes were detected and were lower than the cut-off value established in EFSA guidelines (Additives & Feed, 2012), L. plantarum KU15149 and L. brevis KU15176 were said to be safe. Antibiotic resistance has become a major safety concern for mankind and various organizations, including WHO, U.S. Food and Drug Administration, and EFSA, are raising awareness of this issue.
Whole genome sequencing analysis was conducted to verify the presence of antibiotic resistance genes and virulence genes in the probiotic strain, and the results confirmed the absence of any clinically significant genes. The ResFinder was applied to analyze the presence of antibiotic resistance genes such as ampicillin, vancomycin, kanamycin, streptomycin, erythromycin, clindamycin, tylosin, tetracycline, and chloramphenicol. L. plantarum KU15149 and L. brevis KU15176 both showed no resistance to all above genes as well as by the RGI program. Furthermore, no virulence genes were examined by the genome sequence. According to the report by Wattimury et al. (2023), L. planatrum subsp. plantarum Kita-3 was found to be resistant to clindamycin, streptomycin, and chloramphenicol. Neither of the L. plantarum KU15149 and L. brevis KU15176 used in this study carried antibiotic resistance genes.
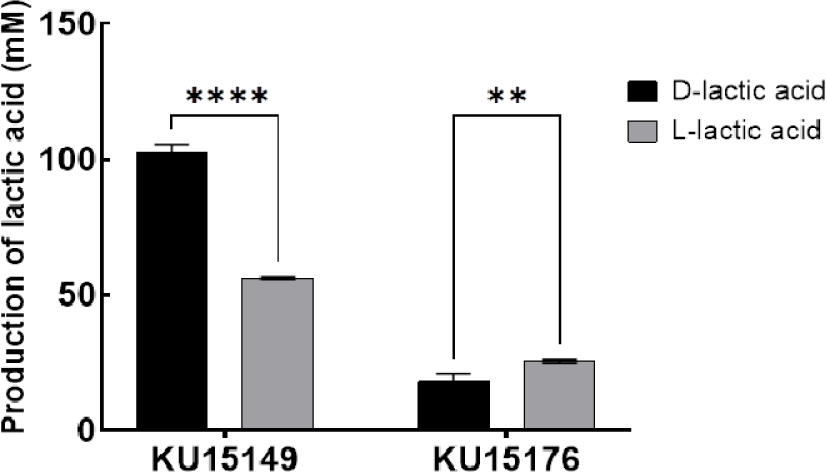
In human body, L-lactate can be metabolized, whereas its isomer D-lactate cannot. Lactic acid bacteria possess DL-lactate racemase, which convert L-lactate to D-lactate. Increased activity of this enzyme may lead to D-lactate accumulation and acidosis, particularly in children and patients with short bowel syndrome (Hove & Mortensen, 1995). In this result, L. plantarum KU15149 produced significantly higher levels of D-lactic acid (102 ppm) compared with L-lactic acid (56.2 ppm). In contrast, L. brevis KU15176 produced significantly lower levels of D-lactic acid (17.9 ppm) compared with L-lactic acid (25.4 ppm) (Fig. 2). L. plantarum has two pathways for the production of D-lactic acid: one via NAD-dependent lactate dehydrogenase, and the other through its incorporation as the terminal residue of the muramoyl-pentadepsipeptide peptidoglycan precursor, which is involved in cell wall biosynthesis. The latter pathway contributes to the natural resistance to vancomycin (Goffin et al., 2005). Although there is no regulation on D-lactic acid, gram-positive anaerobes such as Lactobacillus sp. are known to produce D-lactic acid (Uribarri et al., 1998). On the other hand, L. brevis KU15176, which exhibited low lactic acid production, is consistent with the metabolic characteristics of heterologous fermentation, producing not only lactic acid but also other metabolites such as acetic acid, ethanol, and CO2 (Pérez-Díaz et al., 2017). Therefore, L. brevis KU1517 reflects the strain’s unique metabolic characteristics.
Although the presence of biogenic amines in food may help lactic acid bacteria survive significant amounts of biogenic amines in food are hazardous to humans (Lonvaud-Funel, 2001). Biogenic amines determined in this assay were agmatine, histamine, β- phenylethylamine, putrescine, serotonin, tyramine, tryptamine, and spermidine. Biogenic amine is produced during plants and animals’ metabolism and is a component that is commonly discovered in fermented food (Doeun et al., 2017). In this study, L. plantarum KU15149 was negative for all eight tested biogenic amines, whereas L. brevis KU15176 produced 86.7 ppm of putrescine but was negative for the other seven biogenic amines (data not shown). According to Rauscher-Gabernig et al. (2012), the maximum tolerable concentrations of putrescine in sauerkraut, cheese, and seasoning were reported to be 140, 180, and 510 ppm, respectively. In terms of food safety, the distribution and content of biogenic amines are important, in case of Korea, histamine content is set to 200 mg/kg or less for frozen fish, salted eel, canned food, tyramine and phenylethylamine to 100–800 mg/kg and 30 mg/kg each but specifications for biogenic amines for probiotics have not been established (Arulkumar et al., 2023).
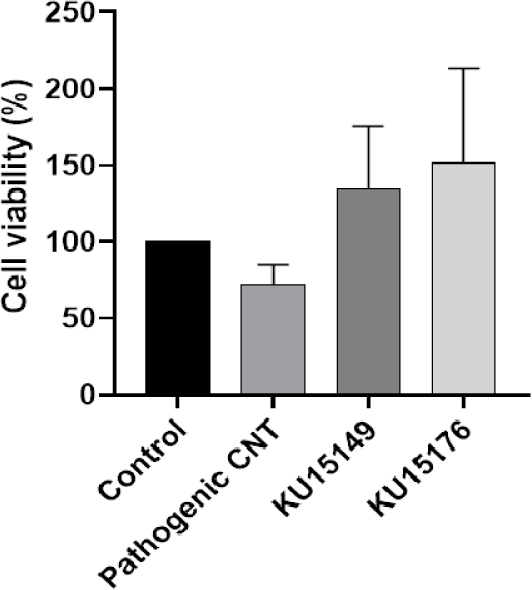
Results were converted to percentages. K. pneumoniae subsp. pneumoniae KCCM 60022 showed cell viability lower than 80% (Fig. 3). Viability of cell lower than 80% with treatment of bacteria, is determined to have a cytotoxicity. Other than the positive strain, the viability of cell was higher than 80% on both of lactic acid bacteria treated. It was reported that the exopolysaccharide of L. plantarum BGAN8 protected Caco-2 cells from cadmium-induced toxicity (Brdarić et al., 2021), and pasteurized L. brevis IBRC- M10790 was effective in preventing inflammation and did not show cytotoxicity at any concentration in the Caco-2 cell line (Ebrahiminejad et al., 2024).
Conclusion
In this study, the safety of L. plantarum KU15149 and L. brevis KU15176 was evaluated through various in vitro assays. Both strains were negative for hemolytic activity, mucin degradation, gelatin liquefaction, urease activity, indole production, β-glucuronidase, and bile salt deconjugation, and also showed acceptable levels in antibiotic susceptibility tests. In the genome analysis, L. plantarum KU15149 was confirmed to harbor the vanX gene; however, this was not considered a safety issue, as lactobacilli are intrinsically resistant to vancomycin. Furthermore, no transferable antibiotic resistance genes or virulence genes were detected. In metabolite analysis, L. plantarum KU15149 produced more D-lactate than L-lactate, while L. brevis KU15176 produced both isomers at similar levels. No biogenic amines were detected in L. plantarum KU15149, and the putrescine identified in L. brevis KU15176 was within an acceptable range. Importantly, neither strain exhibited cytotoxicity in Caco-2 intestinal cells, supporting their potential safety for intestinal application.
